November 25th 2017 - John Guibas & Tejpal Virdi
Abstract
Currently there is strong interest in data-driven approaches to medical image classification. However, medical imaging data is scarce, expensive, and fraught with legal concerns regarding patient privacy. Typical consent forms only allow for patient data to be used in medical journals or education, meaning the majority of medical data is inaccessible for general public research. We propose a novel, two-stage pipeline for generating synthetic medical images from a pair of generative adversarial networks, tested in practice on retinal fundi images. We develop a hierarchical generation process to divide the complex image generation task into two parts: geometry and photorealism. We hope researchers will use our pipeline to bring private medical data into the public domain, sparking growth in imaging tasks that have previously relied on the hand-tuning of models. We have begun this initiative through the development of SynthMed, an online repository for synthetic medical images.
Introduction
Computer-aided medical diagnosis is widely used by medical professionals to assist in the interpretation of medical images. Recently, deep learning algorithms have shown the potential to perform at higher accuracy than professionals in certain medical image understanding tasks, such as segmentation and classification. Along with accuracy, deep learning improves the efficiency ofdata analysis tremendously, due to its automated and computational nature. Since most medical data is produced in large volumes, and is often 3-dimensional (MRIs, CTs, etc.), it can be cumbersome and inefficient to annotate manually.
There is strong interest in computer-aided medical diagnosis systems that rely on deep learning techniques. However, due to proprietary and privacy reasons limiting data access, the development and advancement of these systems cannot be accelerated by public contributions. It is difficult for medical professionals to make most medical images public without patient consent. In addition, the publicly available datasets often lack size and expert annotations, rendering them useless for the training of data-hungry neural networks. The design of these systems is therefore done exclusively by researchers that have access to private data, limiting the growth and potential of this field of research.
In the last 10 years, many breakthroughs in deep learning attribute success to extensive public datasets such as ImageNet. The annual ImageNet competition decreased image recognition error rates from 28.2% to 6.7% in the span of 4 years from 2010 to 2014. ImageNet required the work of almost 50,000 people to evaluate, sort, and annotate one billion candidate images. This showcases that access to large and accurate datasets is extremely important for building accurate models. However, current research in the field of medical imaging has relied on hand-tuning models rather than addressing the underlying problem with data. We believe that a public dataset for medicine can spark exponential growth in imaging tasks.
We propose a novel pipeline for generating synthetic medical images, allowing for the production of a public and extensive dataset, free from privacy concerns. We put this into practice through SynthMed, a public repository for these images.
General Pipeline
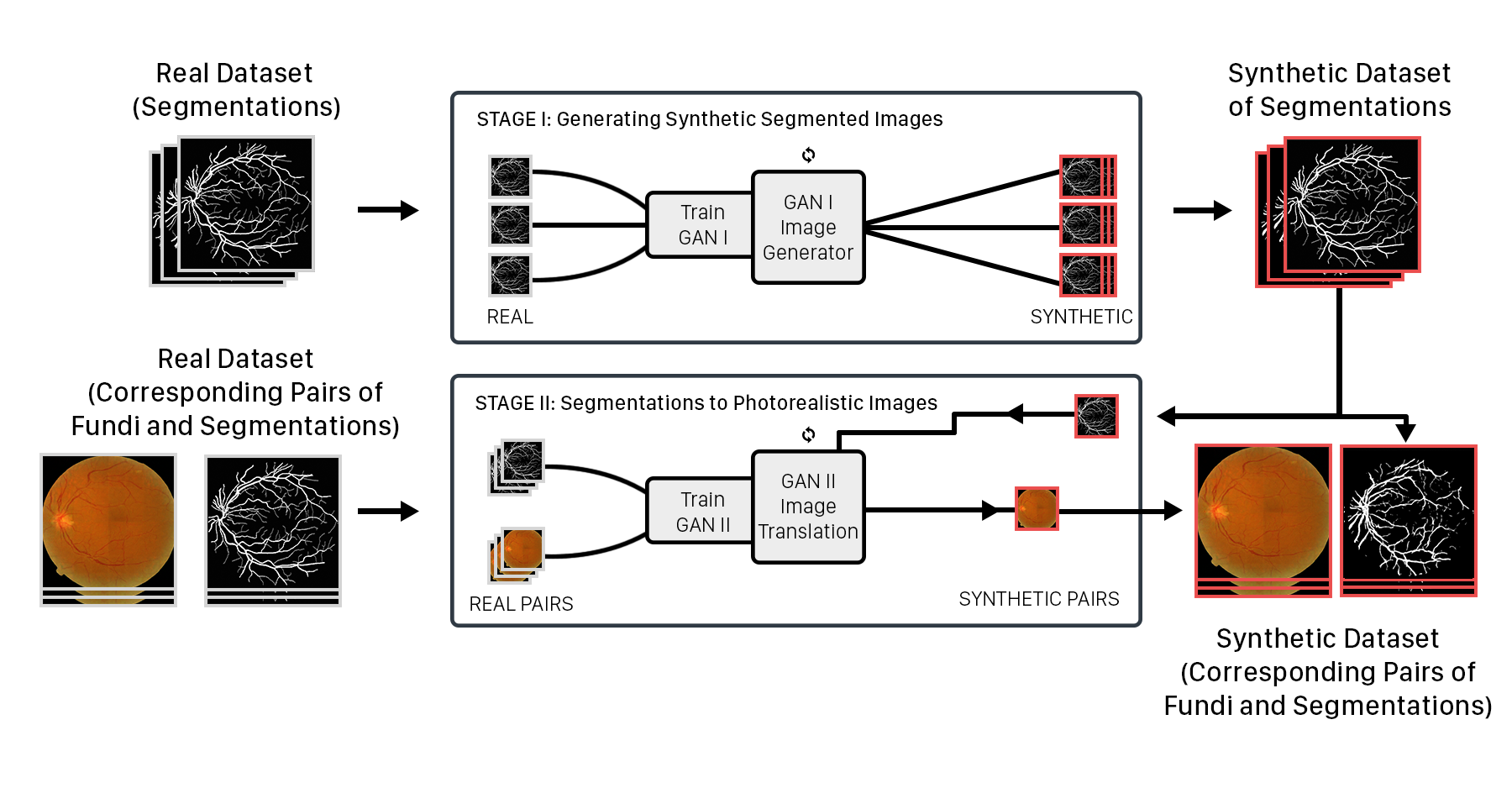
To generate a high quality synthetic dataset, we propose the use of two GANs, breaking down the generation problem into two parts. Stage-I GAN: Produce segmentation masks that represent the variable geometries of the dataset. Stage-II GAN: Translate the masks produced in Stage-I to photorealistic images.
We illustrate the process with retinal fundi images.
Quantitative Results
We received an F1 accuracy rating of 0.8877 for our synthetically trained u-net and an F1 accuracy of 0.8988 for our DRIVE-trained u-net. The negligible difference between the two scores displays the quality of our produced training data.
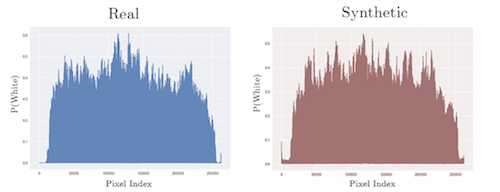
To test for variance, we obtained a KL-divergence score that shows the difference between the distributions of two datasets. The synthetic data score of 4.759 is from comparing the synthetic and real datasets, while the real-data score of 4.212 x 10-4 was measured by comparing two random subsets of the real data. This low score is expected as the two subsets of images are from the same dataset. The synthetic-data score is higher than the real-data score, showing that our synthetic datadoes not simply copy the original distribution.
Discussion
Due to the extreme variation of medical imaging data (various illuminations, noise, patterns, etc.), a single GAN is unable to produce a convincing image. The GAN is unable to determine complex structures, as seen with the poorly defined vessel tree structure and dark spots. It is only able to identify simple features such as general color, shape, and lighting.
This lack of detail is unacceptable for medical image generation, as medical images have many intricacies that must be accurately represented for the data to be usable. Our dual GAN architecture improves the quality of synthetic images by breaking down the challenging task of generating medical images into to hierarchical process. Stacking GANs has been shown to be effective in refining the images produced by a GAN, as seen with Zhang et al. This also allows the unstable nature of GANs to be controlled by providing each GAN with a relatively elementary task. Stage-I GAN focuses only on a much lower dimensional problem: generating unique segmentation geometries, while ignoring photorealism. This allows Stage-II GAN to only generate the colors, lighting, and textures of the medical image from the given geometry. Because the geometry is generated in a lower dimensional image by a separate GAN, an unrealistic vessel geometry causes a larger loss compared to a single GAN that produces unrealistic geometries in its high dimensional fundi images. This system allows both GANs in our pipeline to perform at a high level and reach convergence faster, creating images with more realistic geometries and textures than an ordinary single GAN system.
In addition, the nature of our pipeline produces a wider variety of images than the original dataset. This is because our pipeline generates images that are between the data that formed the distribution. Our synthetic dataset keeps the general statistical distribution of the real dataset while producing original images. Our pipeline can produce larger quantities of images for effective use in data-driven machine learning tasks, while avoiding legal concerns regarding patient privacy.
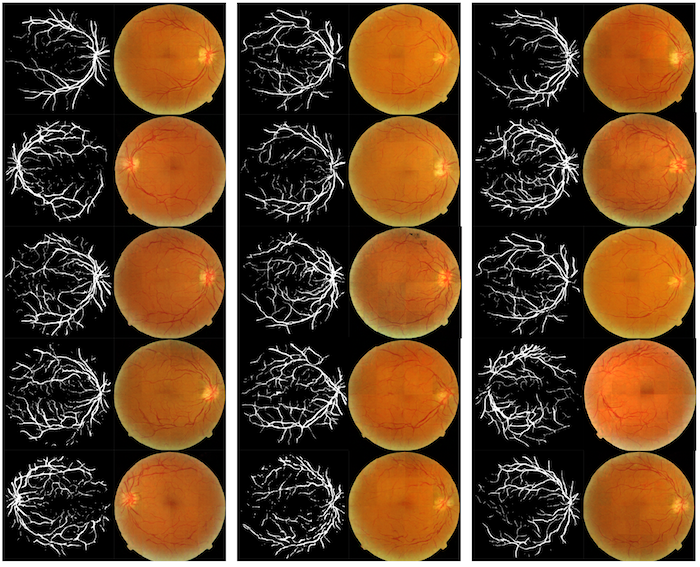
Samples of Generated Images